What we study
Our work utilizes the fascinating characteristics of stem cells to address fundamental questions in cell and cancer biology: How do cells identify, measure, and respond to each other and to their environment? What are the signals that control the renewal and regeneration of tissues? How do these signals go wrong in cancer? Our long-term goal is to uncover an underlying circuit theory behind these behaviors – a set of predictive principles that tell us how complex functionality arises from simpler biological components. We have a particular interest in kinase networks that regulate healthy tissue homeostasis and become damaged in cancer. Through our quantitative high-throughput imaging and drug discovery efforts, we are finding new ways to understand and repair these networks.
Projects
1) Fundamental cell biology of tissue homeostasis and regeneration
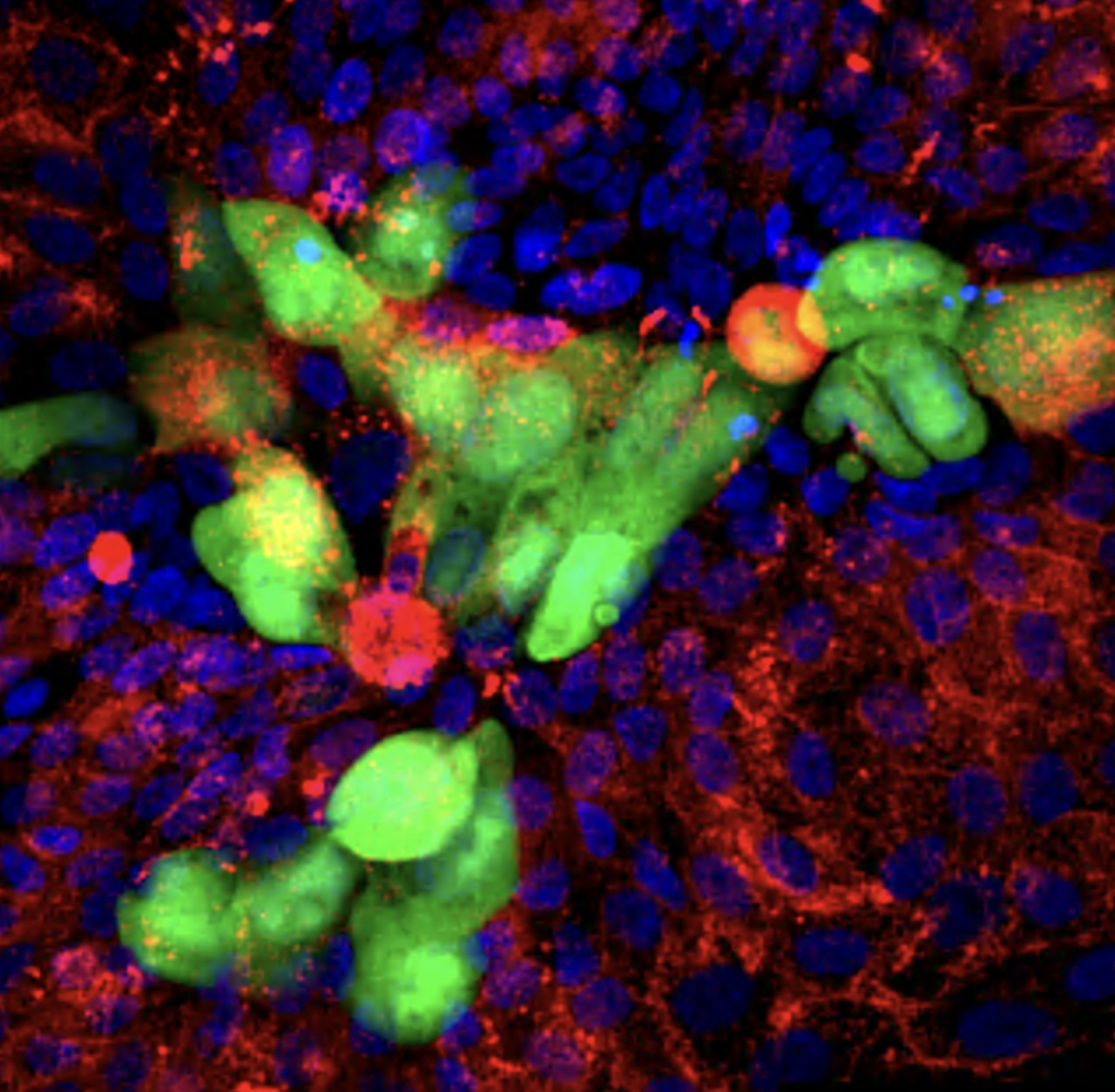
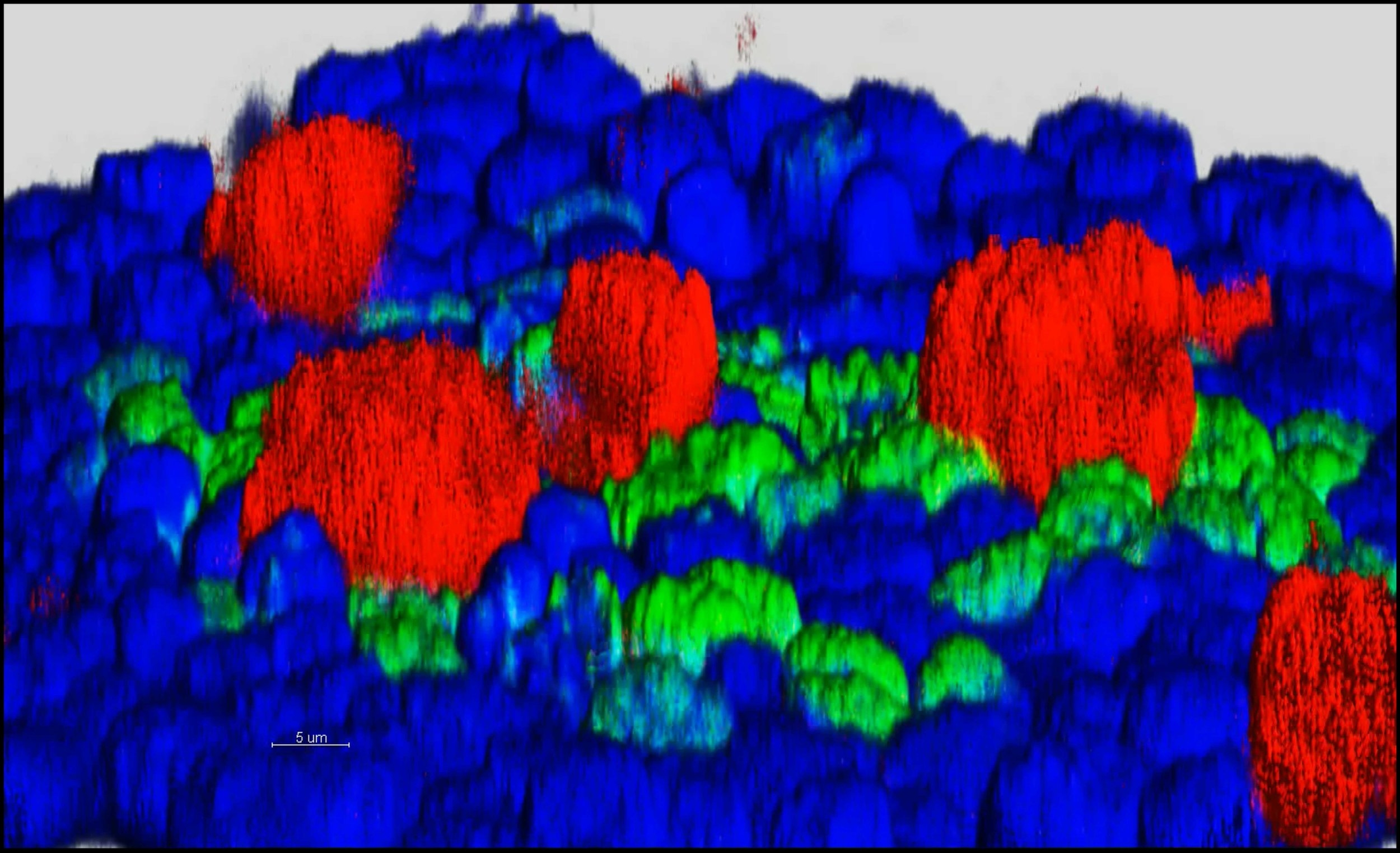
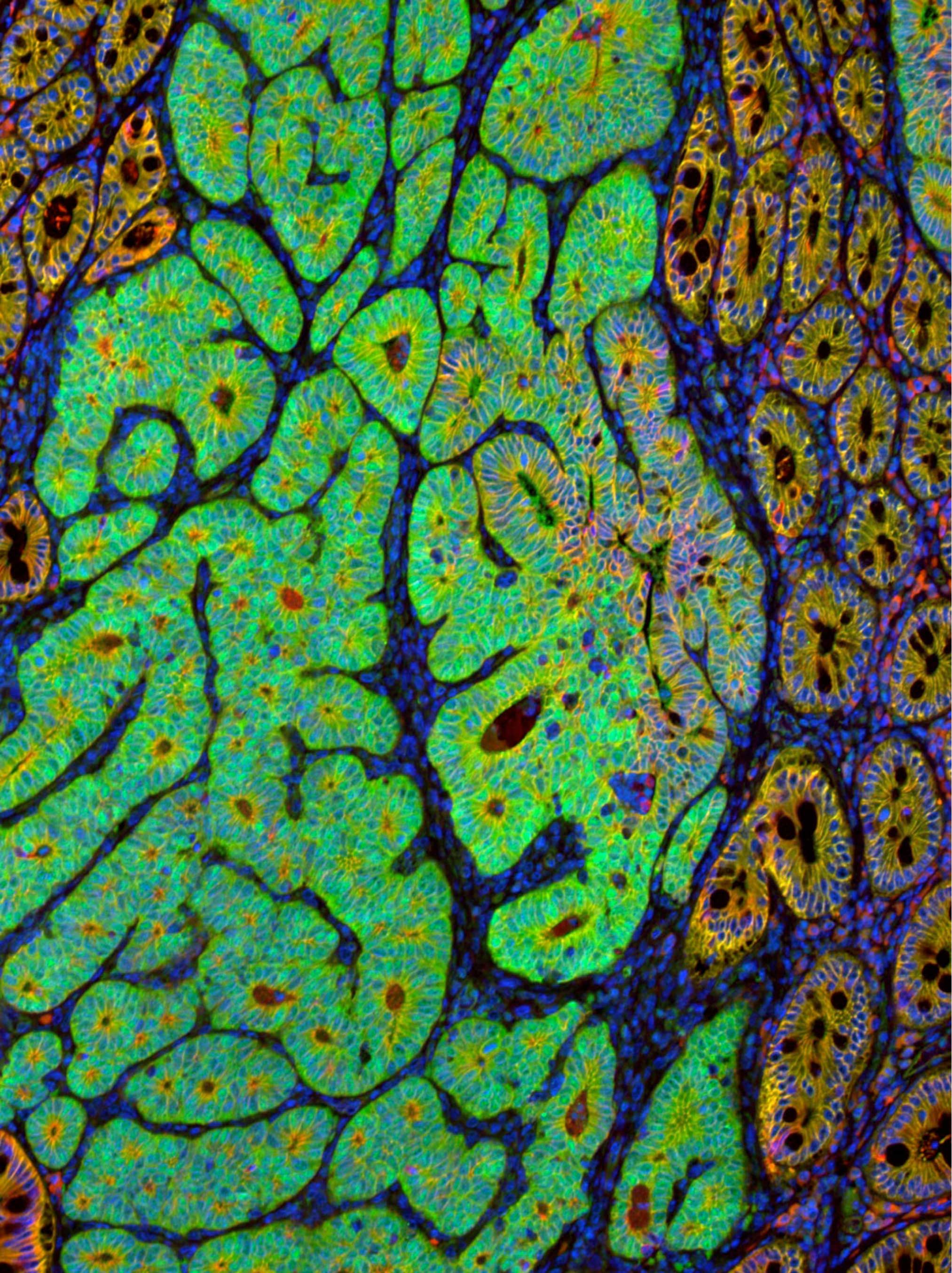
Uncovering new regulators of tissue homeostasis and regeneration
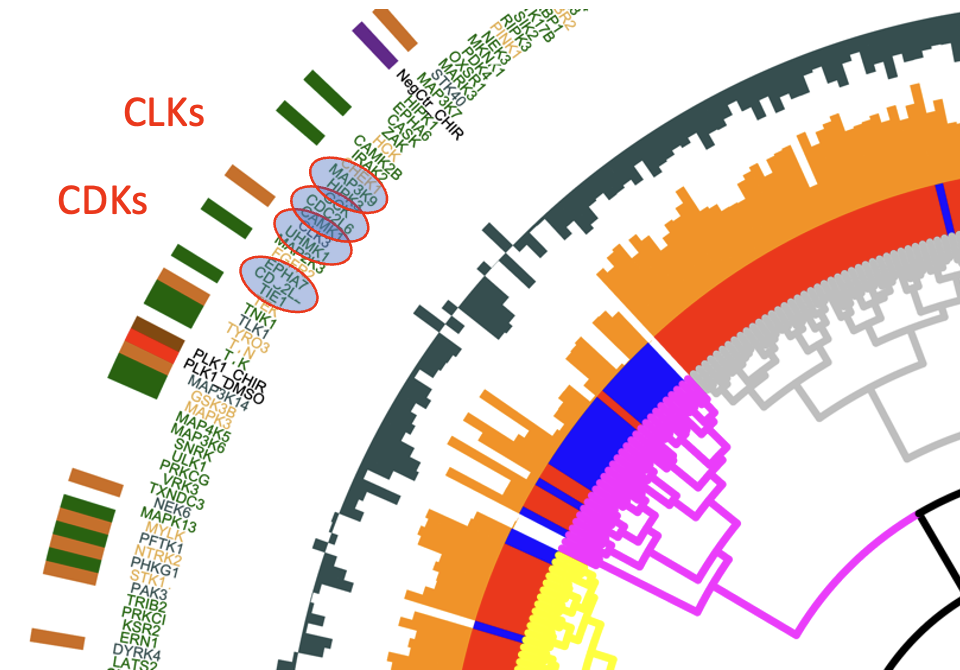
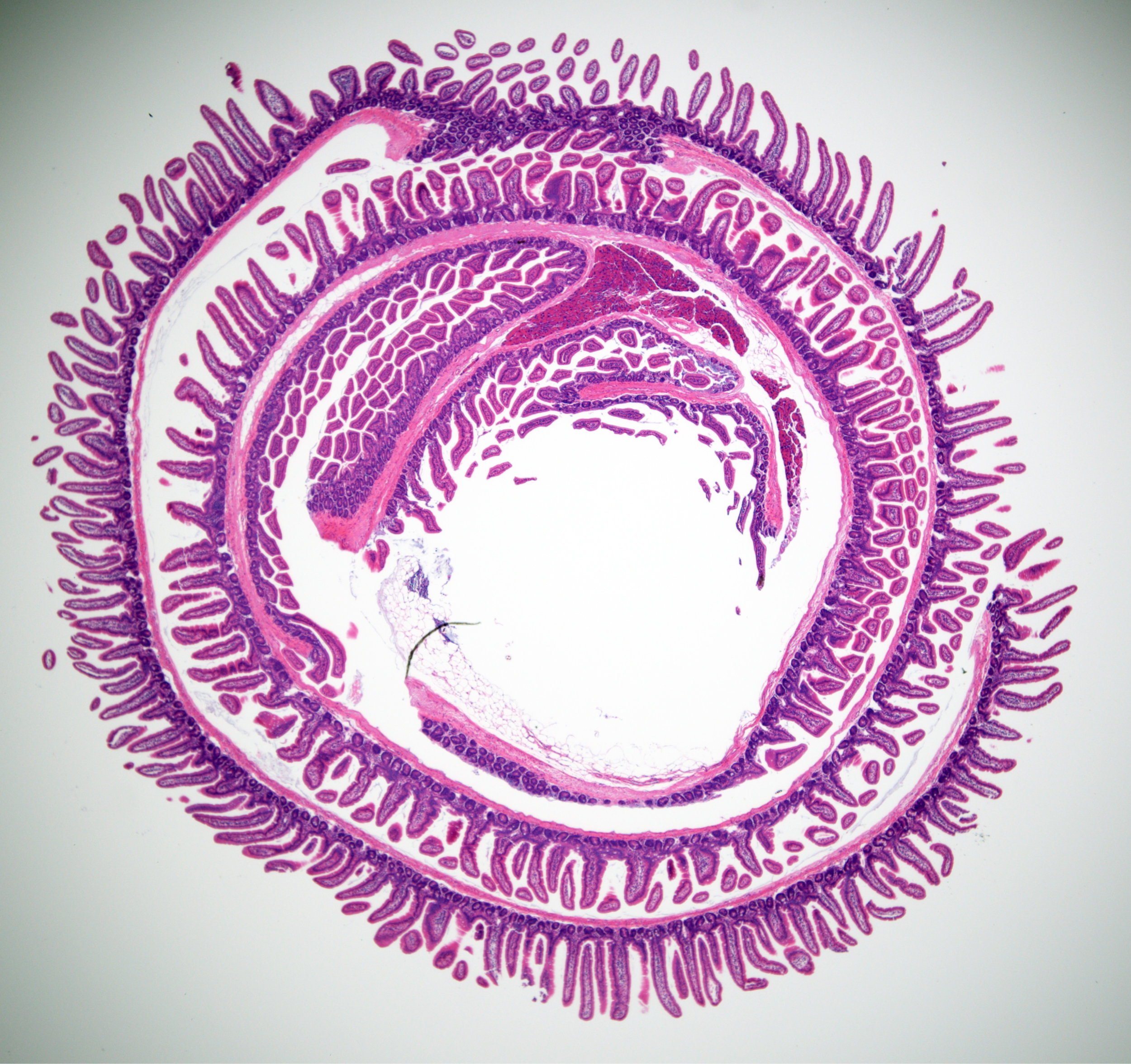
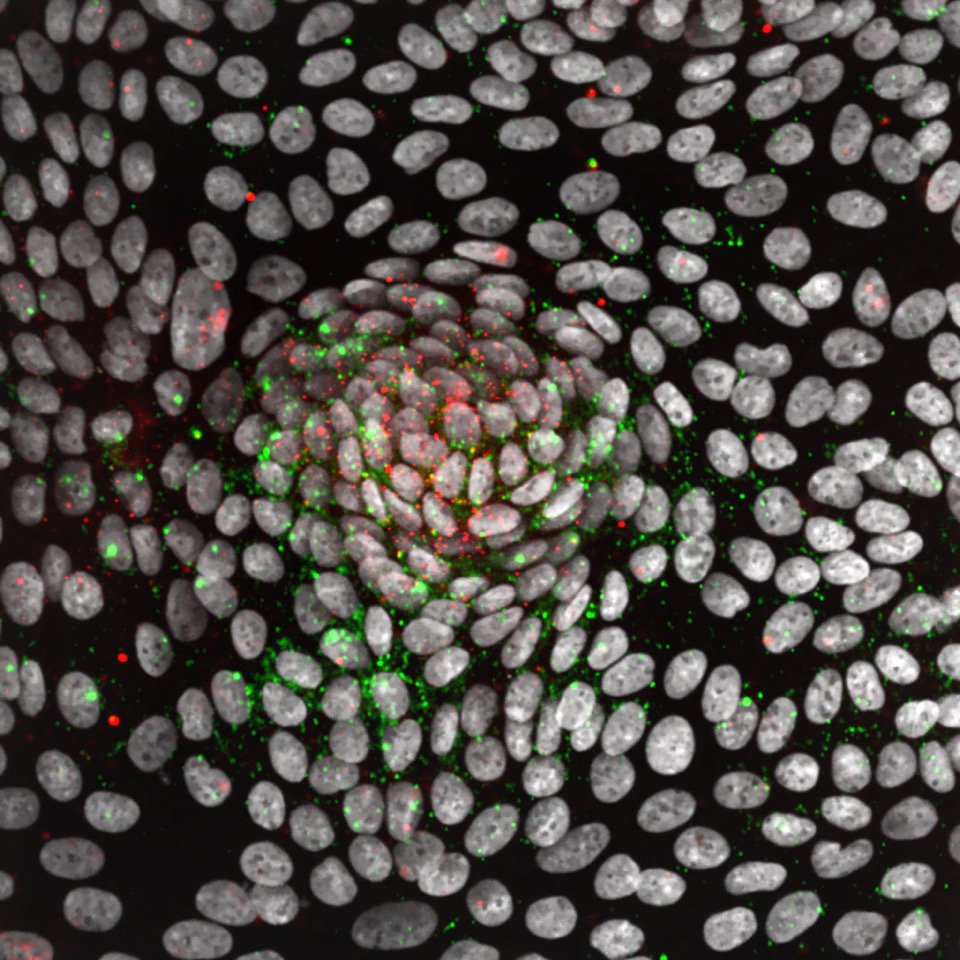
We have established a high-dimensional functional map of kinase activity in colonic stem cells that has uncovered previously unrecognized molecular regulators of intestinal tissue homeostasis, including key members of the CLK and CDK kinase families. This work has defined distinct functional networks and signaling modules that coordinate epithelial maintenance, regeneration, and stress responses, providing a systems-level view of how tissue stability is controlled.
The current phase of the project focuses on validating these kinases, along with other newly identified regulatory players, through targeted functional assays. Using both 2D and 3D intestinal culture systems, we are performing mechanistic studies to define how CLK and CDK signaling influences stem cell behavior, epithelial integrity, and tissue repair. These experiments establish causal roles for these kinases within the broader regulatory networks that maintain intestinal homeostasis.
GSK-3 signaling in crypt maintenance and drug resistance
Evidence suggests the protein kinase, GSK-3, is highly regulated in stem cell niches in vivo. Additionally, our studies have shown decreased GSK-3 activity stimulates stem cell maintenance and provides significant drug resistance to chemotherapies. To understand the role of GSK-3 activity in intestinal crypt homeostasis and drug response, we are developing a single cell live reporter for GSK-3 activity. Using time-lapse microscopy, we are exploring the dynamics of GSK-3 activity in the maintenance of the stem cell niche and identifying microenvironmental signals that regulate GSK-3 and produce drug resistance. The goal is to uncover molecular mechanisms by which epithelia self-organize and evade cytotoxic drugs.
2) Cancer drug discovery
Discovery and characterization of small molecule kinase activators
Small molecule inhibition of kinase activity is well established as a fruitful therapeutic strategy. Yet, widely neglected is the development of small molecule kinase activators for rationally selected targets. GSK-3 is an outstanding candidate for small molecule activation due to its many allosteric pockets and its inactivation in various disease states. We are conducting a two-step (enzymatic followed by cell-based imaging) high throughput screen of novel compound libraries to identify small molecule activators of GSK-3. Utilizing a high-dimensional structure-activity relationship (HD-SAR) approach, we are categorizing the compounds into three major classes: 1) proliferation, 2) metabolism, and 3) cytoskeleton. These compounds will be valuable both as tools for biologists and as potential lead compounds for cancer drug development.
3) Technology development
Organoid-based phenotypic screening platform for infectious diseases
We are developing advanced experimental and computational platforms to study infectious diseases, with particular emphasis on clinically important pathogens such as cytomegalovirus (CMV) and Cryptosporidium. These infections represent major global health challenges, especially for children, immunocompromised individuals, and patients in low- and middle-income countries, where they contribute substantially to morbidity, mortality, and long-term developmental consequences.
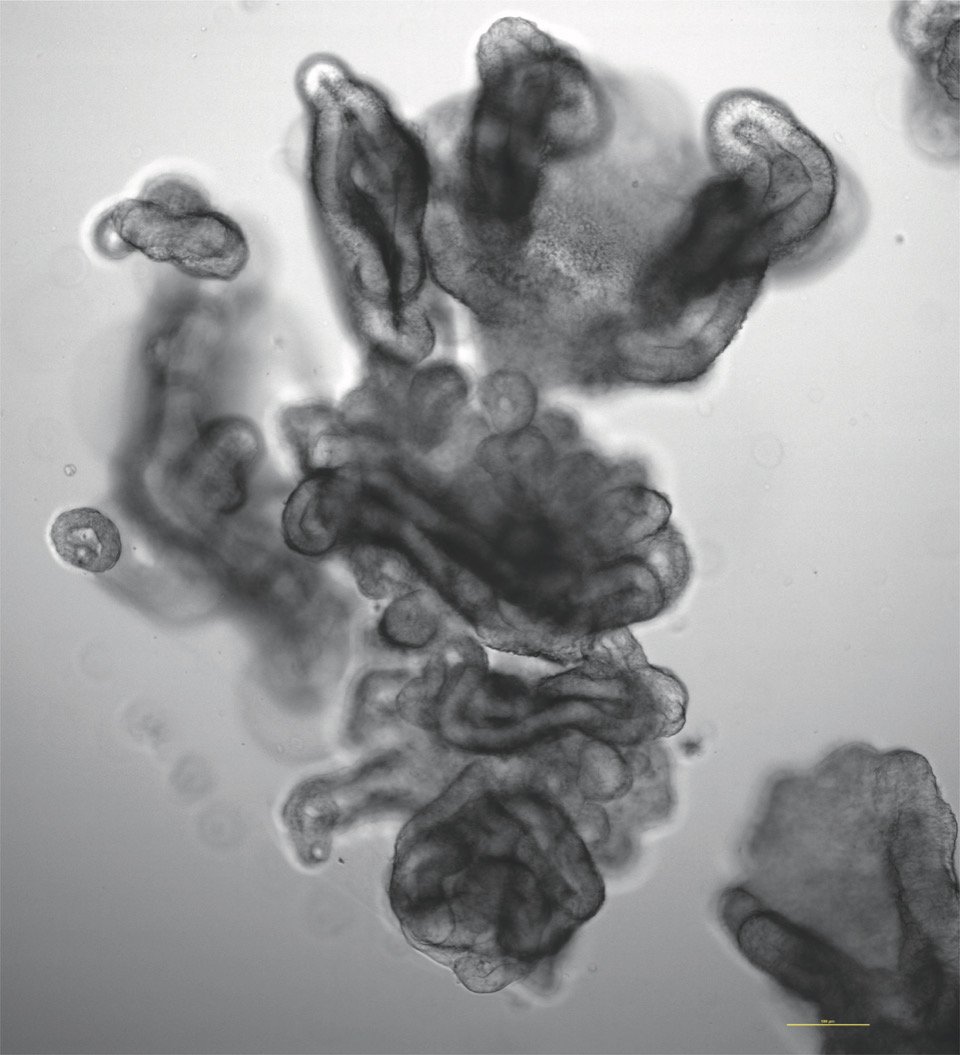
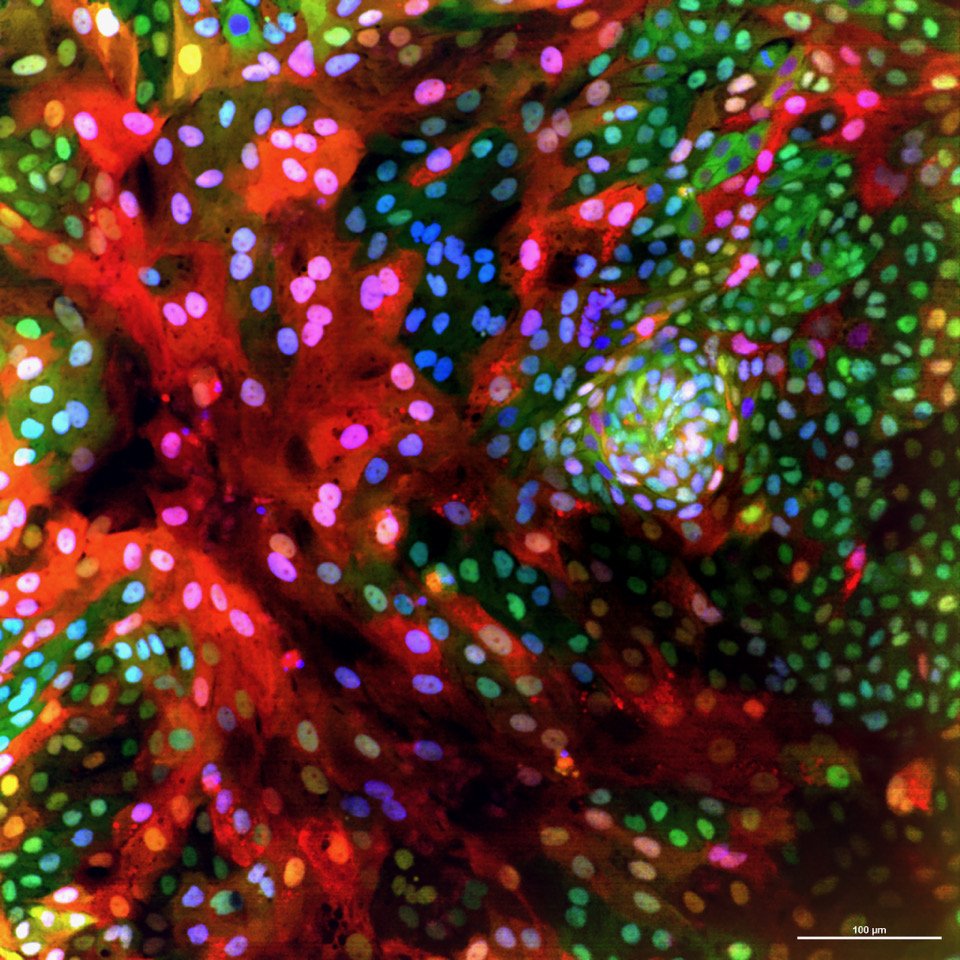
We use human intestinal organoids—“miniature organs” that faithfully recapitulate the structure and biology of the human gut—to model infection in a way that more closely mimics real patient physiology than traditional cell culture systems. Our approach combines large-scale compound testing, high-content microscopy, and artificial intelligence–based image analysis to quantify infection dynamics, host responses, and therapeutic efficacy. By integrating organoid biology with AI-driven phenotypic profiling, we aim to create a broadly applicable pipeline for infectious disease research that can identify and prioritize new therapeutics for pathogens where effective treatments are currently limited or nonexistent.
PEOPLE
Curtis Thorne, PhD
Associate Professor of Cellular and Molecular Medicine and the UA Cancer Center.
Curtis received his B.S. in 2000 from Baylor University, where he concentrated in Biology and Chemistry. Following undergrad, he was a technician for two years at Baylor College of Medicine in the laboratory of Dr. Adrian Lee, studying growth factor signaling in breast cancer. He received his Ph.D. in 2010 from Vanderbilt University in Cell and Developmental Biology under Dr. Ethan Lee. While in graduate school, he discovered a novel therapeutic for the treatment of colon cancer. He conducted postdoctoral studies as an American Cancer Society Fellow at University of Texas Southwestern in the laboratories of Dr. Steven Altschuler and Dr. Lani Wu (now at UCSF). He developed a high throughput method for culturing intestinal stem cells combined with automated cell imaging. Using these approaches, he discovered novel drug combinations for the treatment of colon cancer. While at UT Southwestern, Curtis received an NIH Pathway to Independence Award with additional training in kinase biochemistry in the lab of Dr. Melanie Cobb.
In 2017, Curtis started his independent lab in the Department of Cellular and Molecular Medicine at the University of Arizona. He is also a member of the BIO5 Institute, University of Arizona Cancer Center, and co-founder of two biotech companies, ProxyBio and Branch Therapeutics.
Video and article about Curtis and the Lab.
Email: curtisthorne@email.arizona.edu
Marina Cardó Vila, PhD
Associate Research Professor
Marina received her B.S in 1995 and her M.S in Immunology in 1996 from the University of Barcelona. She received her Ph.D. in 2003 from the University of Barcelona under the mentorship of Dr Antonio Celada (University of Barcelona) and Dr. Renata Pasqualini (The Burnham Institute, La Jolla, CA, USA), studying the biology of macrophages and mechanisms of cellular adhesion. During her postdoctoral fellowship she studied therapeutic applications of tissue-specific protein interactions using in vitro and in vivo phage display in the laboratory of Drs. Wadih Arap and Renata Pasqualini at MD Anderson Cancer Center. In 2013 she joined their team as Assistant Research Professor at the University of New Mexico, where she identified multiple receptor targets in normal and diseased microenvironments, identifying new strategies to mitigate early events of tumor dissemination. Some of the peptides identified in these studies underwent evaluation in a first-in-man phase zero clinical trial. She joined the University of Arizona Cancer Center as Associate Research Professor in 2018 where she studied the regulation of p-body formation and mRNA decapping in breast and prostate cancer in Dr. Andrew Kraft’s lab and drug resistance in castration-resistance prostate cancer in Dr. Cindy Miranti’s Lab. She focused on studying DNA repair in cancer and Lupus disease in Dr. Joann Sweasy’s Lab
Dylan Corcoran
Graduate Student
Dylan received his B.S. in Genetics, Cell Biology, and Development from the University of Minnesota – Twin Cities in 2022. During and after his undergraduate studies, he worked in the lab of Dr. Yasuhiko Kawakami. Dylan studied the zinc finger transcription factor Sall4, a master regulator of vertebral posterior trunk patterning and development. Of note, Dylan and the Kawakami lab established a novel connection between Sall4 and Wnt signaling, which he presented at the 2023 Developmental Biology Center Symposium, an annual international conference hosted by the University of Minnesota. Dylan worked in conjunction with other lab members to publish this finding as part of a larger story on the role of Sall4 in gene regulation and posterior development. He joined the University of Arizona’s ABBS program in 2024 and has decided to pursue his doctorate in cancer biology. Dylan joined the Thorne lab in March of 2025, where he plans to utilize the remarkable model system of intestinal organoids for his future research. In his free time, Dylan enjoys reading, playing/watching sports, and being outdoors.
Briana Guzman
Graduate Student
Briana is a first-generation Hispanic student, born and raised in Tucson. She received her B.S. in Molecular and Cellular Biology from the University of Arizona in 2023. During her undergrad Briana worked in the Nagy Lab investigating the evolution of the gene network that patterns segmentation within the fruit fly Drosophila and the beetle Tribolium. During her time in the lab her interests in research grew, but in fields of infectious diseases, specifically cancer, and working to develop therapeutics in the field. Briana is continuing at the University of Arizona to receive her PhD. In her spare time, Briana enjoys hiking during her free time in the beautiful desert and working out.
Kate Johnson
MD/PhD Student
Kate received her B.S. in Biology-Chemistry from Point Loma Nazarene University in 2022. During undergrad, she worked in the Dorrell-Woelbern Lab, where she studied E7 oncogene variance across HPV subtypes and completed her honors thesis on differentiation of tumor-associated macrophages in the glioblastoma tumor microenvironment. During this time, she also interned at the Lowy Medical Research Institute in San Diego, where she cultured retinal organoids from iPSCs and trained AI to do retinal layer segmentation to help determine the multifactorial pathogenesis of MacTel. When the pandemic began in 2020, Kate also started working both at PLNU’s Wellness Center and in the lab to screen students and staff for COVID-19. Kate joined the MD/PhD program at the University of Arizona in 2022. She officially began her graduate studies with the Thorne Lab in 2024, where she plans to focus on signaling pathways in gut regeneration. In her free time, Kate enjoys watercolor painting, reading, and fostering pets through the Pima Animal Care Center.
Crystal Morales
Graduate Student
Crystal is a proud first-generation student who obtained her bachelor’s in biomedical sciences and Master’s in biology at Northern Arizona University (NAU). In 2020 Crystal was awarded the NIH funded RISE (Research Initiative for Scientific Enhancement) fellowship at NAU. Crystal worked on peptide-based HPV vaccines under Dr. Naomi Lee at NAU and it helped her develop her current research interests which include working on novel cancer therapeutics while also tackling cancer disparities. In 2022, Crystal was awarded the University Fellowship award at the University of Arizona and joined the Arizona Biological and Biomedical Sciences program. Crystal is pursuing her doctorate degree in cancer biology while working in the Thorne lab, where she plans to continue the lab’s research on colorectal cancer. She hopes one day her work can benefit patients and minimize the burden of cancer in marginalized communities. Additionally, Crystal likes to be outdoors, get crafty, and train her puppy, Chimichanga, to do new tricks.
Julia Morris
MD/PhD Student
Julia received her degrees in biology and ethics from Villanova University in 2015. There she worked in the lab of Matthew Youngman, studying immunosenescence, the genetic and biochemical basis of the changes seen in immune response during natural aging. The fall after graduating, she moved from the East Coast out to Arizona to join the MD/PhD Program at U of A. In 2021, she began working in the Thorne Lab for the graduate portion of her dual degree. Julia adores the warm weather of the southwest and spends her free time going to the gym, hiking, cooking, and playing with her three cats, Ellie, Olaf, and Kristoff.
Kelvin Pond, PhD
Assistant Research Professor
Kelvin received his bachelor’s degree in biology in 2010 from the University of Oregon. After this, he obtained his ESL teaching certificate and worked abroad as an English teacher in Prague, CR, and then as a high school science teacher in Bangkok, TH. In 2014, Kelvin moved back to Arizona and received an MS in Biochemistry at Northern Arizona University under Dr. Diane Stearns, studying Uranium Toxicity. Since then, Kelvin has been at the University of Arizona. As a technician, he studied drug resistance mechanisms in pancreatic cancer under Dr. Terry Landowski until joining the lab of Dr. Nathan Ellis in 2015, where he studied DNA repair in human cancers. Kelvin received his PhD in 2019 and is currently working to establish independent projects supported by the Thorne/Paek groups to develop new ways to track signaling states in time and space as tissues regenerate. When not doing experiments, Kelvin basks in the greatest town on the planet, where he enjoys photography, rock climbing, eating tacos, and hanging out with his dog Winston.
Reeba Varghese, PhD
Postdoc
Reeba graduated from Arizona State University with a B.S. in Biological Sciences (Genetics, Cell, and Developmental Biology) in 2014. She then received her Master’s degree in Cellular and Molecular Medicine from the University of Arizona in 2018. As a Master student, Reeba joined Dr. Cynthia Miranti’s lab to investigate the survival pathways induced by various extracellular matrices (i.e., laminin and collagen) in castration-resistant prostate cancer cells that result in drug resistance. In 2023 she finished her doctorate in Cancer Biology at the University of Arizona in the Thorne Lab. Currently, she is postdocing as she continues her drug discovery efforts in the Thorne lab. In her free time, she loves to read, do photography, and watch movies.
Pearl Wichaidit, PhD
Research Scientist IV/Computational Biologist
Pearl received her Ph.D. from Department of Electrical Engineering (Electromagnetics Field & Waves) at University of Wisconsin - Madison. After graduation, she worked on a small project improving computational model for fluorescent light bulb funded by Sylvania as a postdoc. She started her career in Biology as a postdoctoral researcher at Altschuler and Wu laboratory at University of Texas Southwestern Medical Center - Dallas where she met Curtis and found her passion in image and big data analysis. She worked in Melanie Cobb’s laboratory after Dr Altschuler and Dr Wu moved to UCSF before joining Thorne laboratory at 2022.
Lab Alumni
Shravani Daptadar - Master’s Student, currently a researcher at Yale
Carly Cabel - Graduate Student, currently Postdoc at Mayo Clinic, Scottsdale
Hannah Seery - Research Technician
Mireya Pimentel - Undergrad, currently Graduate School at U of Wisconsin Madison
Elaheh Alizadeh - Postdoc, currently Research Scientist, Jax Labs
Analisa Stevens - Undergrad, currently Graduate Student at U of Arizona
Anissa Ferris - Undergrad
Amanda Ruelas - Undergrad, currently Graduate School at Vanderbilt
Julianna Haug - Undergrad, currently Graduate School at Stowers
Jenna Inman - Undergrad, currently Physician Assistant School, Denver
Mary Jo Cantoria - Postdoc, currently Research Scientist, Roche Tissue Diagnostics
David Jones Jr. - Masters Student, currently at Roche Tissue Diagnostics
Taylor Bargenquast - Undergrad
Sylvestor Moses - Residency, Banner University Medical Center
Matthew Estremera - Resarch Assistant
Sofia De La Cruz - Kenyon College
Aditi Nair - Undergrad
Oddities
Pictures and videos of experimentation, simulation, automation and collaboration
Opportunities
Graduate students
We have opportunities for Ph.D. students to train in the Thorne Lab. If you are interested in our research, email Dr. Thorne or drop by and chat!
Postdoctoral applicants
We are always looking for talented and enthusiastic postdocs with strong training as experimentalists or theorists. Candidates with any combination of skills or interests in organoid culture, biochemistry, drug discovery, machine learning, signal processing, or mathematical modeling are encouraged to contact Dr. Thorne.
Check out why we think Tucson and U of A are great places to do science.
Publications
* denotes equal contribution
2026
Schnitkey L, Anthony C, Cardó-Vila M, Loberg MA, Orton G, McCabe A, Trapani J, Rockouski M, Luo J, Chen SC, Ye F, Robbins DJ, Ahmed Y, Doubrovinski K, Weiss VL, Thorne CA*, Lee E*. Wnt signaling is modulated by the buffer-like properties of cadherins. Mol Biol Cell. 2026 Jan 15;:mbcE25040191. doi: 10.1091/mbc.E25-04-0191. [Epub ahead of print] PubMed PMID: 41537764.
2025
Cabel CR, Guzman BA, Alizadeh E, Li S, Holberg C, Wichaidit C, Cusanovich DA, Paek AL, Thatcher GRJ, Van Doorslaer K, Nargi RS, Sutton RE, Suryadevara N, Crowe JE Jr, Carnahan RH, Campos SK, Thorne CA. Cell-based high-content approach for SARS-CoV-2 neutralization identifies unique monoclonal antibodies and PI3K pathway inhibitors. Front Cell Dev Biol. 2025;13:1538934. doi: 10.3389/fcell.2025.1538934. eCollection 2025. PubMed PMID: 40486909; PubMed Central PMCID: PMC12142043.
Kang H, Fitch JC, Varghese RP, Thorne CA, Cusanovich DA. Optimization of a Cas12a-Driven Synthetic Gene Regulatory Network System. ACS Synth Biol. 2025 May 16;14(5):1732-1744. doi: 10.1021/acssynbio.5c00084. Epub 2025 May 2. PubMed PMID: 40316310; PubMed Central PMCID: PMC12091039.
2024
Senthil Kumar S, Gunda V, Reinartz DM, Pond KW, Thorne CA, Santiago Raj PV, Johnson MDL, Wilson JE. Oral streptococci S. anginosus and S. mitis induce distinct morphological, inflammatory, and metabolic signatures in macrophages. Infect Immun. 2024 Mar 12;92(3):e0053623. doi: 10.1128/iai.00536-23. Epub 2024 Jan 30. PubMed PMID: 38289109; PubMed Central PMCID: PMC10929413.
2023
Song H, Sontz RA, Vance MJ, Morris JM, Sheriff S, Zhu S, Duan S, Zeng J, Koeppe E, Pandey R, Thorne CA, Stoffel EM, Merchant JL. High-fat diet plus HNF1A variant promotes polyps by activating β-catenin in early onset colorectal cancer. JCI Insight. 2023 May 23;. doi: 10.1172/jci.insight.167163. [Epub ahead of print] PubMed PMID: 37219942.
Kang H, Fitch JC, Varghese RP, Thorne CA, Cusanovich DA. SGRN: A Cas12a-driven Synthetic Gene Regulatory Network System. bioRxiv. 2023 May 8;. doi: 10.1101/2023.05.08.539911. PubMed PMID: 37214915; PubMed Central PMCID: PMC10197538.
Kassel S, Hanson AJ, Benchabane H, Saito-Diaz K, Cabel CR, Goldsmith L, Taha M, Kanuganti A, Ng VH, Xu G, Ye F, Picker J, Port F, Boutros M, Weiss VL, Robbins DJ, Thorne CA, Ahmed Y, Lee E. USP47 deubiquitylates Groucho/TLE to promote Wnt-β-catenin signaling. Sci Signal. 2023 Feb 7;16(771):eabn8372. doi: 10.1126/scisignal.abn8372. Epub 2023 Feb 7.PMID: 36749823
Lasick KA, Jose E, Samayoa AM, Shanks L, Pond KW, Thorne CA, Paek AL. FOXO Nuclear Shuttling Dynamics are Stimulus Dependent and Correspond with Cell Fate. Mol Biol Cell. 2023 Feb 3:mbcE22050193. doi: 10.1091/mbc.E22-05-0193. PMID: 36735481
Mallick S, Chakrabarti J, Eschbacher J, Moraitis AG, Greenstein AE, Churko J, Pond KW, Livolsi A, Thorne C, Little AS, Yuen KCJ, Zavros Y. Genetically engineered human pituitary corticotroph tumor organoids exhibit divergent responses to glucocorticoid receptor modulators. Transl Res. 2023 Jan 12:S1931-5244(23)00002-6. doi: 10.1016/j.trsl.2023.01.002.
Cantoria MJ*, Alizadeh E*, Ravi J, Varghese RP, Bunnag N, Pond KW, Kettenbach AN, Ahmed Y, Paek AL, Tyson JJ, Doubrovinski K*, Lee E*, Thorne CA*. Feedback in the β-catenin destruction complex imparts bistability and cellular memory. Proc Natl Acad Sci U S A. 2023 Jan 10;120(2):e2208787120. doi: 10.1073/pnas.2208787120. Epub 2023 Jan 4. PubMed PMID: 36598937.
2022
Chakrabarti J, Pandey R, Churko JM, Eschbacher J, Mallick S, Chen Y, Hermes B, Mallick P, Stansfield BN, Pond KW, Thorne CA, Yuen KCJ, Little AS, Zavros Y. Development of Human Pituitary Neuroendocrine Tumor Organoids to Facilitate Effective Targeted Treatments of Cushing's Disease. Cells. 2022 Oct 23;11(21):3344. doi: 10.3390/cells11213344.PMID: 36359740
Pond KW, Morris JM, Alkhimenok O, Varghese RP, Cabel CR, Ellis NA, Chakrabarti J, Zavros Y, Merchant JL, Thorne CA*, Paek AL*.
Live-cell imaging in human colonic monolayers reveals ERK waves limit the stem cell compartment to maintain epithelial homeostasis. Elife. 2022 Sep 12;11:e78837. doi: 10.7554/eLife.78837.
2021
Grant A, Xicola RM, Nguyen V, Lim J, Thorne C, Salhia B, Llor X, Ellis N, Padi M. Molecular drivers of tumor progression in microsatellite stable APC mutation-negative colorectal cancers. Sci Rep. 2021 Dec 6;11(1):23507. doi: 10.1038/s41598-021-02806-x. PubMed PMID: 34873211; PubMed Central PMCID: PMC8648784.
Wu MH, Padilla-Rodriguez M, Blum I, Camenisch A, Figliuolo da Paz V, Ollerton M, Muller J, Momtaz S, Mitchell SAT, Kiela P, Thorne C, Wilson JM, Cox CM. Proliferation in the developing intestine is regulated by the endosomal protein Endotubin. Dev Biol. 2021 Dec;480:50-61. doi: 10.1016/j.ydbio.2021.08.009. Epub 2021 Aug 17. PubMed PMID: 34411593; PubMed Central PMCID: PMC8555764.
Chen EC, Gilchuk P, Zost SJ, Suryadevara N, Winkler ES, Cabel CR, Binshtein E, Chen RE, Sutton RE, Rodriguez J, Day S, Myers L, Trivette A, Williams JK, Davidson E, Li S, Doranz BJ, Campos SK, Carnahan RH, Thorne CA, Diamond MS, Crowe JE Jr. Convergent antibody responses to the SARS-CoV-2 spike protein in convalescent and vaccinated individuals. Cell Rep. 2021 Aug 24;36(8):109604. doi: 10.1016/j.celrep.2021.109604. Epub 2021 Aug 10. PubMed PMID: 34411541; PubMed Central PMCID: PMC8352653.
Tomchaney M, Contoli M, Mayo J, Baraldo S, Li S, Cabel CR, Bull DA, Lick S, Malo J, Knoper S, Kim SS, Tram J, Rojas-Quintero J, Kraft M, Ledford JG, Tesfaigzi Y, Martinez FD, Thorne CA, Kheradmand F, Campos SK, Papi A, Polverino F. Paradoxical effects of cigarette smoke and COPD on SARS-CoV-2 infection and disease. BMC Pulm Med. 2021 Aug 23;21(1):275. doi: 10.1186/s12890-021-01639-8. PubMed PMID: 34425811; PubMed Central PMCID: PMC8381712.
Perez-Miller S, Patek M, Moutal A, Duran P, Cabel CR, Thorne CA, Campos SK, Khanna R. Novel Compounds Targeting Neuropilin Receptor 1 with Potential To Interfere with SARS-CoV-2 Virus Entry. ACS Chem Neurosci. 2021 Apr 21;12(8):1299-1312. doi: 10.1021/acschemneuro.0c00619. Epub 2021 Mar 31. PubMed PMID: 33787218; PubMed Central PMCID: PMC8029449.
2020
Pond KW, Doubrovinski K, Thorne CA. Wnt/β-catenin Signaling in Tissue Self-Organization. Genes (Basel). 2020 Aug 14;11(8). doi: 10.3390/genes11080939. Review. PubMed PMID: 32823838; PubMed Central PMCID: PMC7464740.
Sanman LE, Chen IW, Bieber JM, Thorne CA*, Wu LF*, Altschuler SJ*. Generation and Quantitative Imaging of Enteroid Monolayers. Methods Mol Biol. 2020;2171:99-113. doi: 10.1007/978-1-0716-0747-3_6. PubMed PMID: 32705637.
2019
Cabel CR, Alizadeh E, Robbins DJ, Ahmed Y, Lee E*, Thorne CA*. Single-Cell Analyses Confirm the Critical Role of LRP6 for Wnt Signaling in APC-Deficient Cells. Dev Cell. 2019 Jun 17;49(6):827-828. doi: 10.1016/j.devcel.2019.05.039. PubMed PMID: 31211991; PubMed Central PMCID: PMC6613640.
2018
Miyata N, Morris LL, Chen Q, Thorne C, Singla A, Zhu W, Winter M, Melton SD, Li H, Sifuentes-Dominguez L, Llano E, Huff-Hardy K, Starokadomskyy P, Lopez A, Reese TA, Turer E, Billadeau DD, Winter SE, Burstein E. Microbial Sensing by Intestinal Myeloid Cells Controls Carcinogenesis and Epithelial Differentiation. Cell Rep. 2018 Aug 28;24(9):2342-2355. doi: 10.1016/j.celrep.2018.07.066. PubMed PMID: 30157428; PubMed Central PMCID: PMC6177233.
Thorne CA*, Chen IW*, Sanman LE, Cobb MH, Wu LF, Altschuler SJ. Enteroid Monolayers Reveal an Autonomous WNT and BMP Circuit Controlling Intestinal Epithelial Growth and Organization. Dev Cell. 2018 Mar 12;44(5):624-633.e4. doi: 10.1016/j.devcel.2018.01.024. Epub 2018 Mar 1. PubMed PMID: 29503158; PubMed Central PMCID: PMC5849535.
Accompanying Preview.
2017
Coster, A. D.*, C. A. Thorne*, L. F. Wu, and S. J. Altschuler, "Examining Crosstalk among Transforming Growth Factor β, Bone Morphogenetic Protein, and Wnt Pathways.", J Biol Chem, vol. 292, issue 1, pp. 244-250, 2017 Jan 06. PMCID: PMC5217683 PMID: 27895117
Coate, K. C., G. Hernandez, C. A. Thorne, S. Sun, T. D. V. Le, K. Vale, S. A. Kliewer, and D. J. Mangelsdorf, "FGF21 Is an Exocrine Pancreas Secretagogue.", Cell Metab, vol. 25, issue 2, pp. 472-480, 2017 Feb 07. PMCID: PMC5299054 PMID: 28089565
Li, B., D. Orton, L. R. Neitzel, L. Astudillo, C. Shen, J. Long, X. Chen, K. C. Kirkbride, T. Doundoulakis, M. L. Guerra, J. Zaias, D. L. Fei, J. Rodriguez-Blanco, C. Thorne, Z. Wang, K. Jin, D. M. Nguyen, L. R. Sands, F. Marchetti, M. T. Abreu, M. H. Cobb, A. J. Capobianco, E. Lee, D. J. Robbins, "Differential abundance of CK1α provides selectivity for pharmacological CK1α activators to target WNT-dependent tumors.", Sci Signal, vol. 10, issue 485, 2017 Jun 27. PMID: 28655862
2016
Wang, Z., O. Tacchelly-Benites, E. Yang, C. A. Thorne, H. Nojima, E. Lee, and Y. Ahmed, "Wnt/Wingless Pathway Activation Is Promoted by a Critical Threshold of Axin Maintained by the Tumor Suppressor APC and the ADP-Ribose Polymerase Tankyrase.", Genetics, vol. 203, issue 1, pp. 269-81, 2016 05. PMCID: PMC4858779 PMID: 26975665
2015
Thorne, C. A.*, C. Wichaidit*, A. D. Coster, B. A. Posner, L. F. Wu, and S. J. Altschuler, "GSK-3 modulates cellular responses to a broad spectrum of kinase inhibitors.", Nat Chem Biol, vol. 11, issue 1, pp. 58-63, 2015 Jan. PMCID: PMC4270937 PMID: 25402767
Karra, A. S., C. A. Taylor, C. A. Thorne, and M. H. Cobb, "A Kinase Divided.", Cancer Cell, vol. 28, issue 2, pp. 145-7, 2015 Aug 10.PMID: 26267529
2013
Saito-Diaz, K., T. W. Chen, X. Wang, C. A. Thorne, H. A. Wallace, A. Page-McCaw, and E. Lee, "The way Wnt works: components and mechanism.", Growth Factors, vol. 31, issue 1, pp. 1-31, 2013 Feb. PMCID: PMC3697919 PMID: 23256519
Hao, J., A. Ao, L. Zhou, C. K. Murphy, A. Y. Frist, J. J. Keel, C. A. Thorne, K. Kim, E. Lee, and C. C. Hong, "Selective small molecule targeting β-catenin function discovered by in vivo chemical genetic screen.", Cell Rep, vol. 4, issue 5, pp. 898-904, 2013 Sep 12.PMCID: PMC3923627 PMID: 24012757
2012
Hang, B. I.*, C. A. Thorne*, D. J. Robbins, S. S. Huppert, L. A. Lee, and E. Lee, "Screening for small molecule inhibitors of embryonic pathways: sometimes you gotta crack a few eggs.", Bioorg Med Chem, vol. 20, issue 6, pp. 1869-77, 2012 Mar 15. PMCID: PMC3298638 PMID: 22261025
2011
Thorne, C. A., B. LaFleur, M. Lewis, A. J. Hanson, K. K. Jernigan, D. C. Weaver, K. A. Huppert, T. W. Chen, C. Wichaidit, C. S. Cselenyi, et al., "A biochemical screen for identification of small-molecule regulators of the Wnt pathway using Xenopus egg extracts.", J Biomol Screen, vol. 16, issue 9, pp. 995-1006, 2011 Oct. PMCID: PMC3694444 PMID: 21859680
Ni, T. T., E. J. Rellinger, A. Mukherjee, S. Xie, L. Stephens, C. A. Thorne, K. Kim, J. Hu, E. Lee, L. Marnett, et al., "Discovering small molecules that promote cardiomyocyte generation by modulating Wnt signaling.", Chem Biol, vol. 18, issue 12, pp. 1658-68, 2011 Dec 23. PMCID: PMC3645312 PMID: 22195568
2010
Jernigan, K. K., C. S. Cselenyi, C. A. Thorne, A. J. Hanson, E. Tahinci, N. Hajicek, W. M. Oldham, L. A. Lee, H. E. Hamm, J. R. Hepler, et al., "Gbetagamma activates GSK3 to promote LRP6-mediated beta-catenin transcriptional activity.", Sci Signal, vol. 3, issue 121, pp. ra37, 2010 May 11. PMCID: PMC3088111 PMID: 20460648
Thorne, C. A., A. J. Hanson, J. Schneider, E. Tahinci, D. Orton, C. S. Cselenyi, K. K. Jernigan, K. C. Meyers, B. I. Hang, A. G. Waterson, et al., "Small-molecule inhibition of Wnt signaling through activation of casein kinase 1α.", Nat Chem Biol, vol. 6, issue 11, pp. 829-36, 2010 Nov. PMCID: PMC3681608 PMID: 20890287
Alfaro, M. P., A. Vincent, S. Saraswati, C. A. Thorne, C. C. Hong, E. Lee, and P. P. Young, "sFRP2 suppression of bone morphogenic protein (BMP) and Wnt signaling mediates mesenchymal stem cell (MSC) self-renewal promoting engraftment and myocardial repair.", J Biol Chem, vol. 285, issue 46, pp. 35645-53, 2010 Nov 12. PMCID: PMC2975189 PMID: 20826809
Saraswati, S., M. P. Alfaro, C. A. Thorne, J. Atkinson, E. Lee, and P. P. Young, "Pyrvinium, a potent small molecule Wnt inhibitor, promotes wound repair and post-MI cardiac remodeling.", PLoS One, vol. 5, issue 11, pp. e15521, 2010 Nov 29. PMCID: PMC2993965 PMID: 21170416
2008
Cselenyi, C. S., K. K. Jernigan, E. Tahinci, C. A. Thorne, L. A. Lee, and E. Lee, "LRP6 transduces a canonical Wnt signal independently of Axin degradation by inhibiting GSK3's phosphorylation of beta-catenin.", Proc Natl Acad Sci U S A, vol. 105, issue 23, pp. 8032-7, 2008 Jun 10. PMCID: PMC2430354 PMID: 18509060
2007
Tahinci, E., C. A. Thorne, J. L. Franklin, A. Salic, K. M. Christian, L. A. Lee, R. J. Coffey, and E. Lee, "Lrp6 is required for convergent extension during Xenopus gastrulation.", Development, vol. 134, issue 22, pp. 4095-106, 2007 Nov. PMCID: PMC4428168 PMID: 17965054
2003
Thorne, C., and A. V. Lee, "Cross talk between estrogen receptor and IGF signaling in normal mammary gland development and breast cancer.", Breast Dis, vol. 17, pp. 105-14, 2003. PMID: 15687681

Contact Info
Leon Levy Building, UA Cancer Center
1515 N. Campbell Ave.
Office: Room 4947
Office phone: 520-626-0395
Lab: Room 4906
Lab phone: 520-626-3265
Tucson, AZ 85724